Abstract
Introduction: Despite the major advances in hematopoietic stem cell transplant (HSCT), there is significant need for better and more objective means for the diagnosis and prediction of acute graft-vs.-host disease (aGVHD). Most of the studies in predicting GVHD focused on the donor cells rather than on the host microenvironment. Using expression data collected by targeted RNA next generation sequencing (NGS) of bone marrow samples, we explored the potential of RNA data using machine learning to predict aGVHD and overall survival.
Methods: RNA was extracted from bone marrow aspiration samples collected from 98 patients who received HSCT transplant. This included patients with AML (#27), MDS (#37), ALL (#9), CML (#3), MPN (#15), T-cell lymphoma (6), and severe aplastic anemia (SAA) (#1). Post-transplant samples were available on only 85 patients while pre-transplant samples were available on all 98 patients. Females were 50.5%. The average age was 61 years (range: 30 to 75). Pre-transplant samples were collected within 90 days of transplantation; post-transplant samples were collected within 90 days after transplantation. aGVHD was diagnosed and staged per MAGIC criteria. The expression levels of 1408 genes were quantified using NGS. This panel of genes included inflammatory and oncogenic genes. We developed a machine learning algorithm that first selected the relative genes based on the performance of each gene with cross-validation and based on stability measures using statistical significance tests. The selected genes were then used to predict aGVHD or survival with k-fold cross-validation procedure (k=12). A naïve Bayesian classifier was constructed on the training of k-1 subsets and tested on the other testing subset. We applied geometric mean naïve Bayesian (GMNB) as the classifier for prediction.
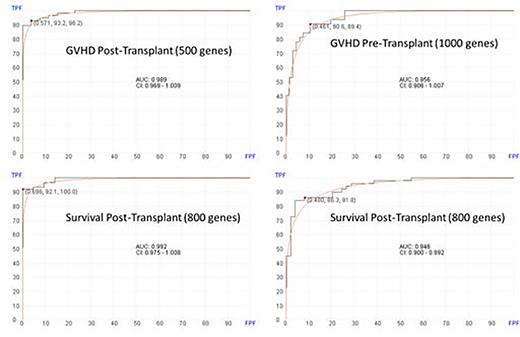
Results: Of the 98 transplanted patients, 66 (67%) developed aGVHD of any stage. Using post-transplant bone marrow RNA data, the machine learning algorithm selected 500 genes for predicting aGVHD with AUC of 0.989 (95% CI: 0.969-1.00), 93.2% sensitivity and 96.2% specificity. Using pre-transplant bone marrow RNA profiles, the machine learning algorithm selected 1000 genes for predicting GVHD with AUC of 0.956 (95% CI: 0.906-1.00), sensitivity of 90.6% and specificity of 89.4%. Of the 85 patients with post-transplant samples, 77 (90.5%) were alive at a median of 15 months at the time of data analysis. Using the post-transplant sample, the algorithm predicted survival using 800 genes with AUC of 0.992 (95% CI : 0.992-1.00), sensitivity of 92.1% and specificity of 100%. Using pre-transplant samples, the algorithm predicted survival using 800 genes with AUC of 0.946 (95% CI: 0.900-0.992), sensitivity of 86.3% and specificity of 91.8%.
Conclusion: Targeted transcriptome of bone marrow samples can predict aGVHD and overall survival with relatively high accuracy when a large number of genes is used. Although the accuracy of this prediction is higher when post-transplant transcriptomic data is used, pre-transplant bone marrow transcriptomic data remains very reliable in such predictions. This confirms that the host bone marrow microenvironment may play a significant role in the development of GVHD and in overall survival in patients undergoing HSCT transplant.
Disclosures
Albitar:Genomic Testing Cooperative: Current Employment, Current holder of stock options in a privately-held company. Ip:AstraZeneca: Membership on an entity's Board of Directors or advisory committees; SecuraBio: Membership on an entity's Board of Directors or advisory committees; Seagen: Speakers Bureau; Pfizer: Honoraria; TG Therapeutics: Membership on an entity's Board of Directors or advisory committees. Goy:Incyte: Honoraria, Other: Steering Committee, Research Funding; Bristol Meyers Squibb: Honoraria, Other: Scientific Advisory Board, Research Funding; Celgene: Consultancy, Honoraria, Research Funding; Elsevier Practice Update: Oncology: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees; AstraZeneca: Honoraria, Other: Steering Committee, Research Funding; Resilience: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Kite: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; AbbVie: Consultancy; MorphoSys: Honoraria, Other: Steering Committee, Research Funding; Physicians’ Education Resource: Consultancy, Honoraria; Vincerx: Honoraria, Other: Scientific Advisory Board; Xcenda: Consultancy, Honoraria; OncLive Peer Exchange: Consultancy, Honoraria; Pharmacyclics: Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Steering committee, Research Funding; Medscape: Consultancy, Honoraria; Michael J. Hennessy Associates, Inc.: Consultancy, Honoraria; Acerta: Research Funding; Genentech: Research Funding; Hoffmann-La Roche: Research Funding; Clinical Advances in Hematology & Oncology: Consultancy, Honoraria; lloplex: Current holder of stock options in a privately-held company, Honoraria, Membership on an entity's Board of Directors or advisory committees; Karyopharm: Research Funding; Infinity Pharmaceuticals: Research Funding; Rosewell Park: Consultancy, Honoraria; Janssen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: Steering Committee, Research Funding; Novartis: Consultancy, Honoraria; Genomic Testing Cooperative: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Cancer Outcome Tracking Analysis: Current equity holder in private company, Membership on an entity's Board of Directors or advisory committees; Seattle Genetics: Research Funding; Verastem: Research Funding; OMI: Current Employment; Regional Cancer Care Associates: Current Employment. Rowley:SIRPant Immunotherapeutics: Consultancy; ReAlta Life Sciences: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal